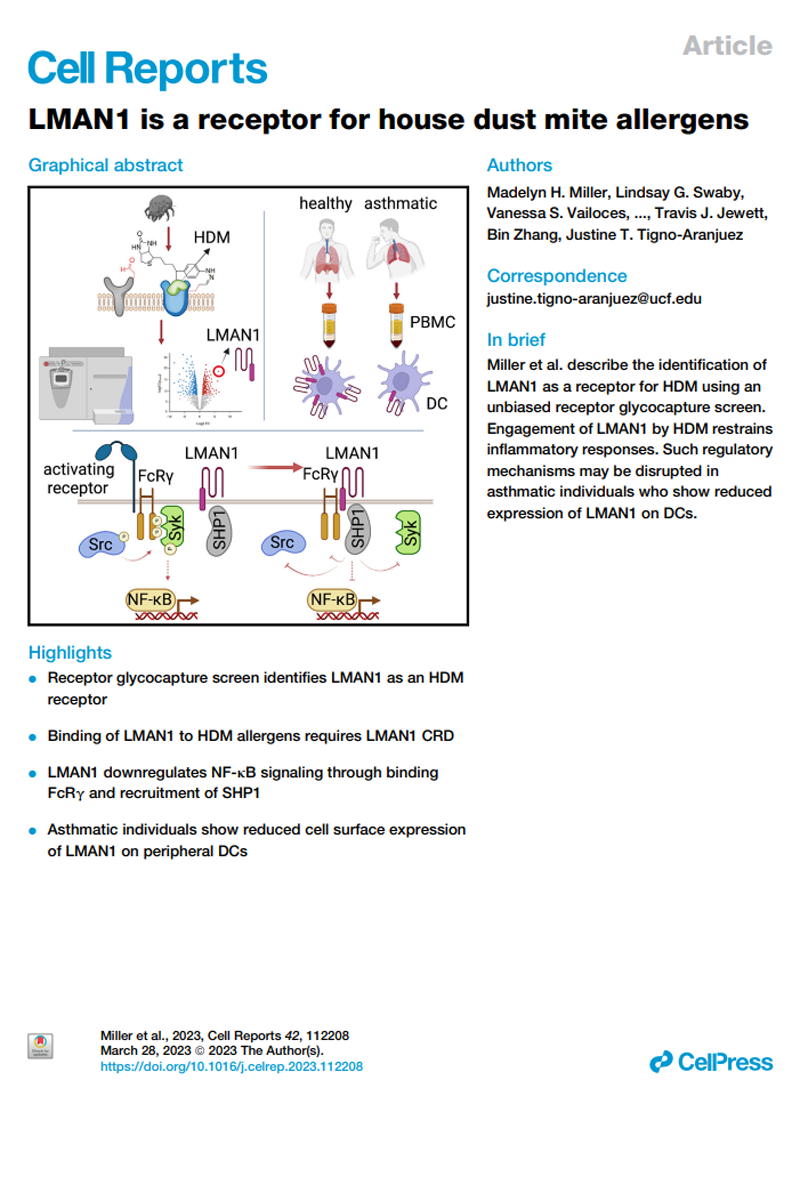
Identification of a receptor for house dust mite allergens
LRC-TriCEPS identified a completely new mechanism of monocyte activation that might be clinically relevant in Alzheimers’ and inflammation diseases. Feb 2023
Sensei Biotherapeutics Presents Preclinical Data at the 37th Society for Immunotherapy of Cancer (SITC) Annual Meeting
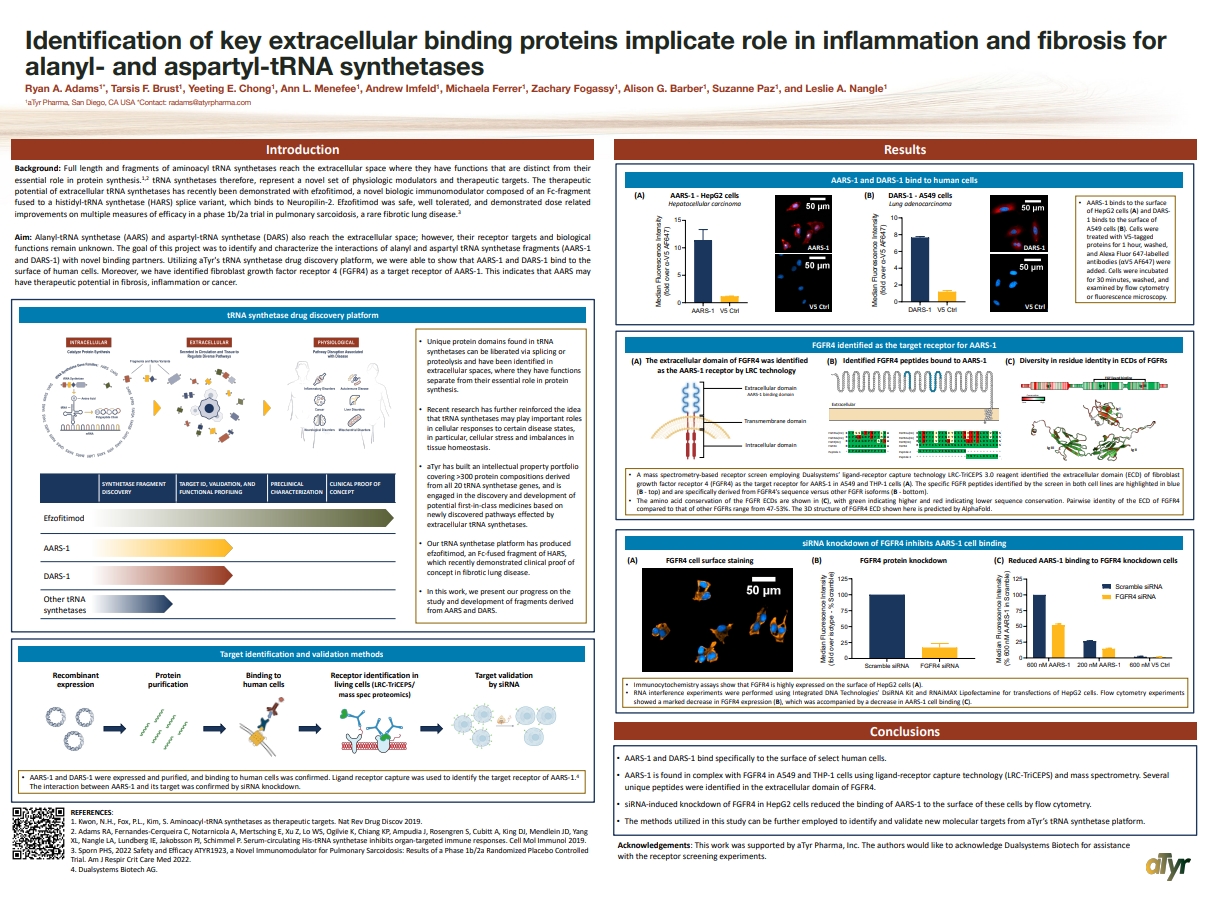
Identification of key extracellular binding proteins
Identification of key extracellular binding proteins implicate role in inflammation and fibrosis for alanyl- and aspartyl-tRNA synthetases
Akkermansia muciniphila secretes a gluc.-like peptide-1-inducing protein that improves glucose homeostasis and ameliorates metabolic disease in mice
Nature Microbiol (2021) https://doi.org/10.1038/s41564-021-00880-5
Neuroplastin Modulates Anti-inflammatory Effects of MANF
iScience Volume 23, ISSUE 12, 101810, December 18, 2020, DOI:https://doi.org/10.1016/j.isci.2020.101810
NKG2A/CD94 Is a New Immune Receptor for HLA-G and Distinguishes Amino Acid Differences in the HLA-G Heavy Chain
International Journal of Molecular Sciences. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7352787/
Acidity changes immunology: a new VISTA pathway
Nature Article VISTA interaction with PSGL-1 identified by LRC-TriCEPS
https://www.nature.com/articles/s41586-019-1674-5 Nature volume 574, pages565–570 (2019)
Anti-VISTA antibody that inhibits Vista function and blocks interaction with PSGL-1 and VSIG3 proteins slows tumor growth
https://www.nature.com/articles/s41598-020-71519-4 Mehta, N., Maddineni, S., Kelly, R.L. et al. An engineered antibody binds a distinct epitope and is a potent inhibitor of murine and human VISTA. Sci Rep 10, 15171 (2020). https://doi.org/10.1038/s41598-020-71519-4 PDF
Phage resistance at the cost of virulence
Phage resistance at the cost of virulence: Listeria monocytogenes serovar 4b requires galactosylated teichoic acids for InlBmediated invasion
PLOS Pathogens | https://doi.org/10.1371/journal.ppat.1008032 October 7, 2019
PDF
TFF3 interacts with LINGO2 to regulate EGFR activation for protection against colitis and gastrointestinal helminths
Nature Communications volume 10, Article number: 4408 (2019) https://doi.org/10.1038/s41467-019-12315-1
White Paper Ligand-Receptor Identification Methodologies Details Matter
European Biopharmaceutical Review April 2019, pages 16-20 / Maria P. Pavlou and Paul Helbling / White -Paper PDF
Validation of extracellular ligand–receptor interactions by Flow‑TriCEPS
BMC Research Notes 2018 11:863 https://doi.org/10.1186/s13104-018-3974-5 Laura A. Lopez‑Garcia, Levent Demiray, Sandra Ruch‑Marder, Ann‑Katrin Hopp, Michael O. Hottiger, Paul M. Helbling and Maria P. Pavlou Received: 28 October 2018 – Accepted: 30 November 2018- Published: 5 December 2018 PDF
Cardiac Targeting Peptide, a Novel Cardiac Vector: Studies in Bio-Distribution, Imaging Application, and Mechanism of Transduction
Maliha Zahid, Kyle S. Feldman, Gabriel Garcia-Borrero, Timothy N. Feinstein, Nicholas Pogodzinski, Xinxiu Xu, Raymond Yurko, Michael Czachowski , Yijen L. Wu, Neale S. Mason and CeciliaW. Lo Biomolecules 2018, 8, 147; doi:10.3390/biom8040147 Received: 24 September 2018 / Accepted: 8 November 2018 / Published: 14 November 2018
Leukocyte differentiation by histidine-rich glycoprotein/stanniocalcin-2 complex regulates murine glioma growth through modulation of anti-tumor immunity
Francis P Roche, Ilkka Pietilä, Hiroshi Kaito, Elisabet O Sjöström, Nadine Sobotzki, Oriol Noguer, Tor Persson Skare, Magnus Essand, Bernd Wollscheid, Michael Welsh and Lena Claesson-Welsh DOI: 10.1158/1535-7163.MCT-18-0097 Received January 27, 2018, Revision received April 21, 2018, Accepted June 19, 2018, Copyright ©2018, American Association for Cancer Research. PDF
Glycomics and Proteomics Approaches to Investigate Early Adenovirus–Host Cell Interactions
Lisa Lasswitz, Naresh Chandra, Niklas Arnberg, Gisa Gerold jmb Journal of Molecular Biology, doi.org/10.1016/j.jmb.2018.04.039 Received 15 February 2018, Revised 24 April 2018, Accepted 30 April 2018, Available online 7 May 2018.
HATRIC-based identification of receptors for orphan ligands
Nadine Sobotzki, Michael A. Schafroth, Alina Rudnicka, Anika Koetemann, Florian Marty, Sandra Goetze, Yohei Yamauchi, Erick M. Carreira & Bernd Wollscheid Nature Communications, volume 9, Article number: 1519 (2018) doi:10.1038/s41467-018-03936-z Published online: 17 April 2018
Staphylococcal Superantigens Use LAMA2 as a Coreceptor GPCT signaling To Activate T Cells
Zhigang Li, Joseph J. Zeppa, Mark A. Hancock, John K. McCormick, Terence M. Doherty, Geoffrey N. Hendy and Joaquín Madrenas J Immunol January 15, 2018, ji1701212; DOI: https://doi.org/10.4049/jimmunol.1701212 (Published online February 5, 2018) This work was supported by the Canadian Institutes for Health Research. J.M. holds a tier I Canada Research Chair in Human Immunology. The Department of Microbiology and Immunology Flow Cytometry and Cell Sorting Facility and McGill Surface Plasmon Resonance–Mass Spectrometry Facility are supported by the Canada Foundation for Innovation.
Toll like receptors TLR1/2, TLR6 and MUC5B as binding interaction partners with cytostatic proline rich polypeptide 1 in human chondrosarcoma
International Journal of Oncology, published online on: November 9, 2017 doi.org/10.3892/ijo.2017.4199 Authors: Karina Galoian, Silva Abrahamyan, Gor Chailyan, Amir Qureshi, Parthik Patel, Gil Metser, Alexandra Moran, Inesa Sahakyan, Narine Tumasyan, Albert Lee, Tigran Davtyan, Samvel Chailyan and Armen Galoyan Metastatic chondrosarcoma is a bone malignancy not responsive to conventional therapies; new approaches and therapies are urgently needed.
Phenotypic screening—the fast track to novel antibody discovery
ScienceDirekt, doi.org/10.1016/j.ddtec.2017.03.004 Department of Antibody Discovery and Protein Engineering, MedImmune, Milstein Building, Granta Park, Cambridge CB21 6GH, UK Available online 25 April 2017
Identification of Putative Receptors for the Novel Adipokine CTRP3 Using Ligand-Receptor Capture Technology
PLoS One. 2016 Oct 11;11(10):e0164593. doi: 10.1371/journal.pone.0164593. eCollection 2016. Li Y1, Ozment T2, Wright GL1, Peterson JM1,3. We used Ligand-receptor glycocapture technology with TriCEPS-based ligand-receptor capture (LRC-TriCEPS; Dualsystems Biotech AG). The LRC-TriCEPS experiment with CTRP3-FLAG protein as ligand and insulin as a control ligand was performed on the H4IIE rat hepatoma cell line.