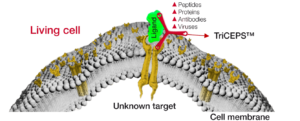
Flow-TriCEPS
for flow cytometry
Cellular responses to ligands such as peptides, proteins, pharmaceutical drugs or entire pathogens are generally mediated through interactions with proteins expressed at the cell surface. The LRC-TriCEPS kit is designed to directly identify the target proteins of your ligand on the living cells directly using near-physiological conditions.
Dualsystems now offers the pretest TriCEPS experiments as services or as a kit.
The pretest test is designed to screen different cell types and decide which cells to use later for the identification of the unknown targets of your ligand of interest in the LRC-TriCEPS main experiment.
In case functional assays are available TriCEPS coupled ligands can also be tested to see if a similar output can be achieved with a TriCEPS coupled ligand compared to a ligand that is not coupled to TriCEPS.
As first step, the TriCEPS v.2.0 molecule is coupled to the ligand of interest (peptide, protein, Antibody, ADC or other primary amine containing molecules) and to the positive control ligand (e.g. transferrin) and negative control ligand (e.g. glycine). This coupling reaction is tested with Dot blot to assess if the coupling worked.
Then, the TriCEPS coupled ligands are added to the non-oxidized cells to assess whether the ligand binds to the unknown targets at the cell surface and TriCEPS does not interfere with the ligand receptor interaction.
Flow Cytometry tests with TriCEPS coupled ligand on oxidized cells can be further performed to test cell viability and ligand target binding as it will occur in the main LRC-TriCEPS experiment.
After successful completion of all pretests, the main experiment can be planned and carried out as described in the LRC-TriCEPS user manual choosing one of the service options provided by Dualsystems Biotech AG.
Contact form
Please fill out all mandatory (*) fields.
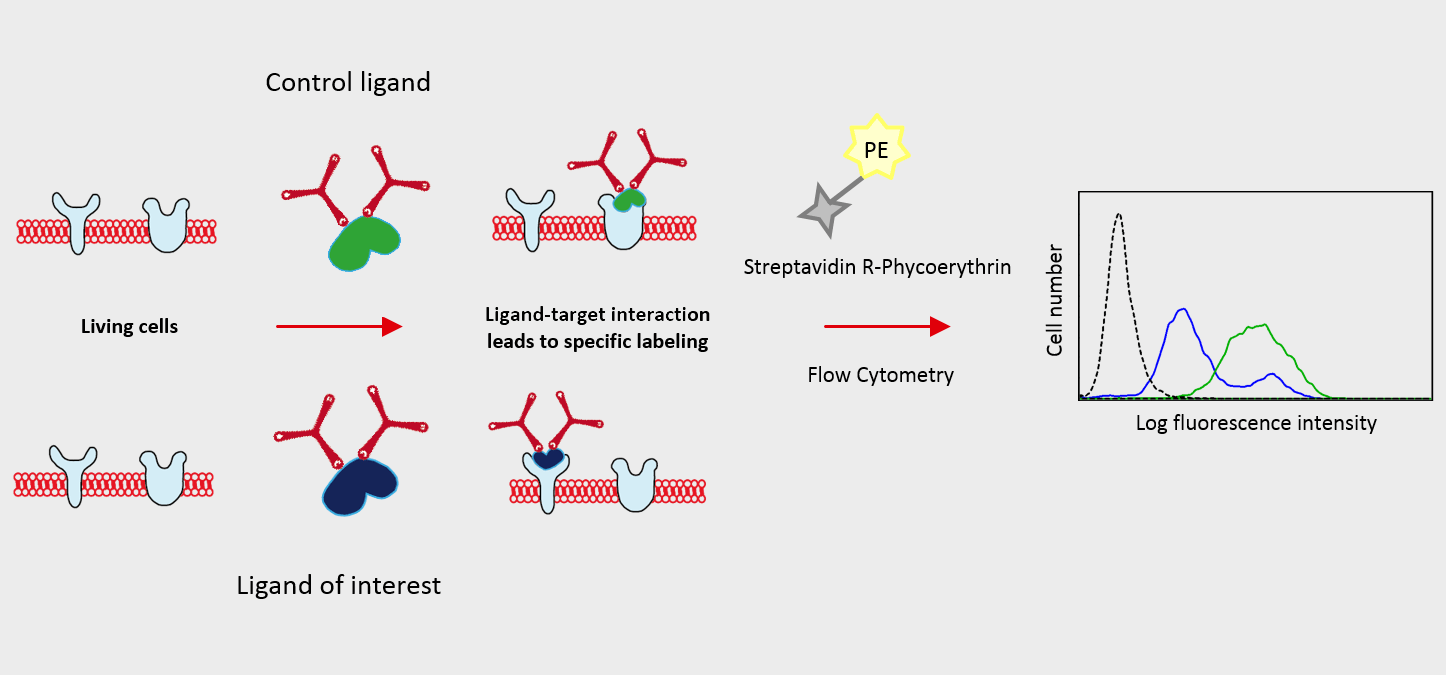
Using Flow-TriCEPS with flow cytometry
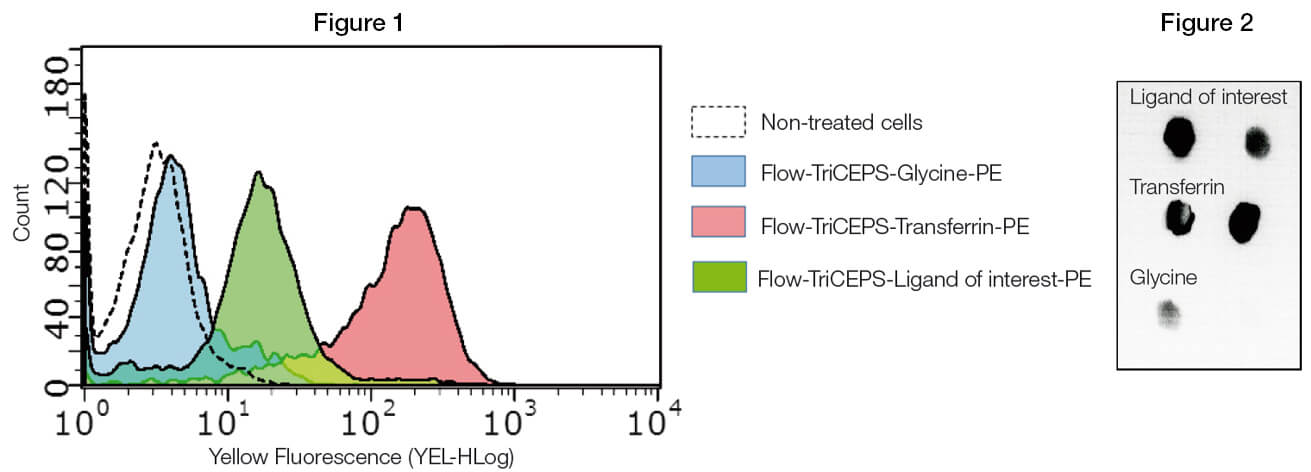
Ligand of interest and controls are coupled to TriCEPS. Coupling of TriCEPS to the ligand can be controlled by dot blot for protein ligands. Cells expressing the unknown targets are treated with TriCEPS coupled ligand and binding is visualized using a fluorophore.
Example of a Flow-TriCEPS experiment
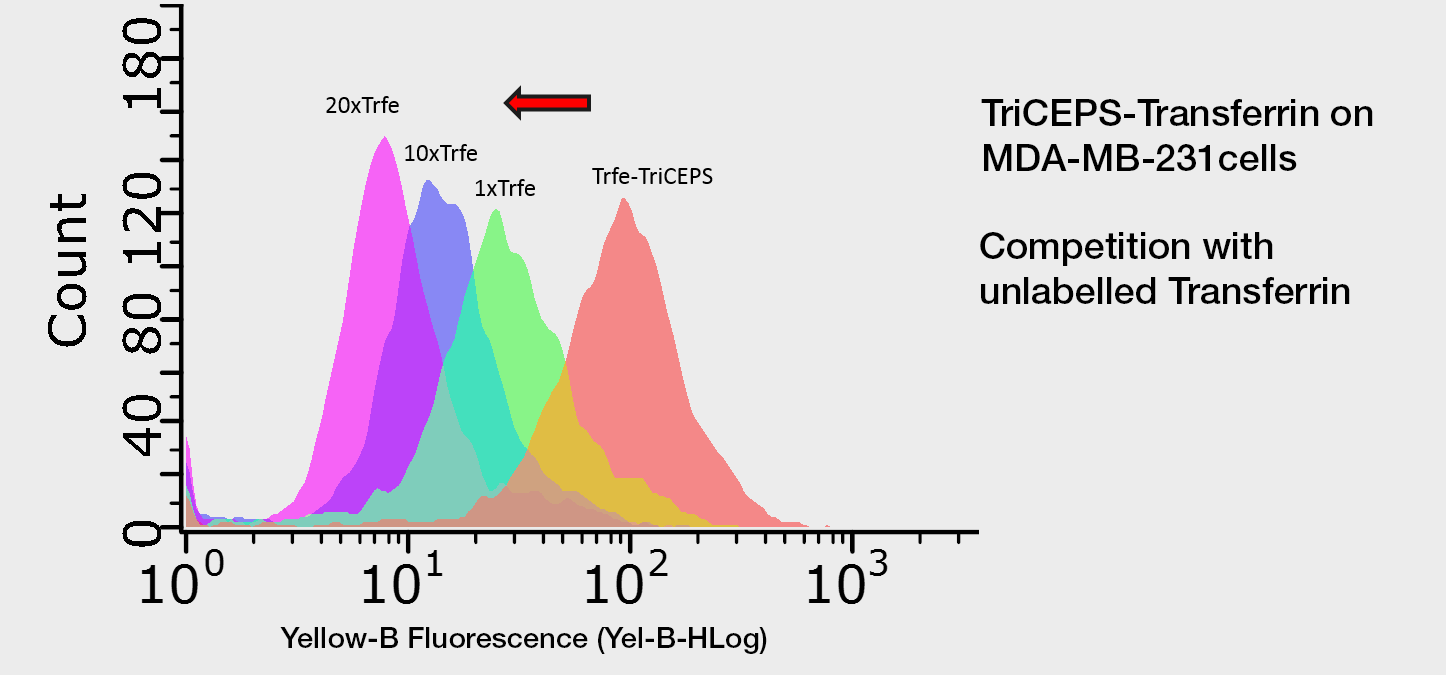
Competition experiment using Flow-TriCEPS. MDA-MB-231 cells are treated with the TriCEPS coupled ligand (orange). In order to confirm that the observed binding is due to the binding of transferrin to its target on the cells, cells are treated with different amounts of unlabeled transferrin before adding TriCEPS coupled ligand. The more unlabeled transferrin is added the smaller the shift of TriCEPS-transferrin is observed since the unlabeled transferrin competes with the TriCEPS coupled transferrin for the binding to its receptor.
TriCEPS-Transferrin on MDA cells
Flow TriCEPS-Kit flyer
Ligand-receptor capture LRC-TriCEPSflyer
(Figure 1) Flow-TriCEPS is coupled to transferrin (positive control), glycine (negative control) and the ligand of interest and added on MDA-MB-231 breast cancer cells. Coupling efficiency is controlled by dot blot (Figure 2). The mean fluorescent shift is compared across the different ligands. It is expected the ligand of interest gives a larger shift than glycine. If different parameters are tested (time, temperature, pH, etc.) the condition leading to the largest shift for the ligand of interest is used for the main identification experiment.
Identify the mode of action
using the TriCEPS platforms
Customers Testimonials – LRC-TriCEPS Service
Testimonials from our customers who have used the LRC-TriCEPS technology – in collaboration with Dualsystems Biotech AG.
LRC-TriCEPS customers worldwide
Over 200 satisfied customers from 28 countries.
LRC-TriCEPS publications worldwide
LRC-TriCEPS / HATRIC-LRC Publications
Concerning the LRC-TriCEPS or HATRIC-LRC platforms.
Use of LRC-TriCEPS to characterize the surfaceome of extracellular vesicles (EV)
Presented at the annual meeting of the International Society for Extracellular Vesicles (ISEV) 2025 Annual Meeting in April
The LRC-TriCEPS platform identified 2 new receptors for ApoA-1, potentially new drug targets for Diabetes Type 2
Apolipoprotein A-I priming via SR-BI and ABCA1 receptor binding upregulates mitochondrial metabolism to promote insulin secretion in INS-1E cells
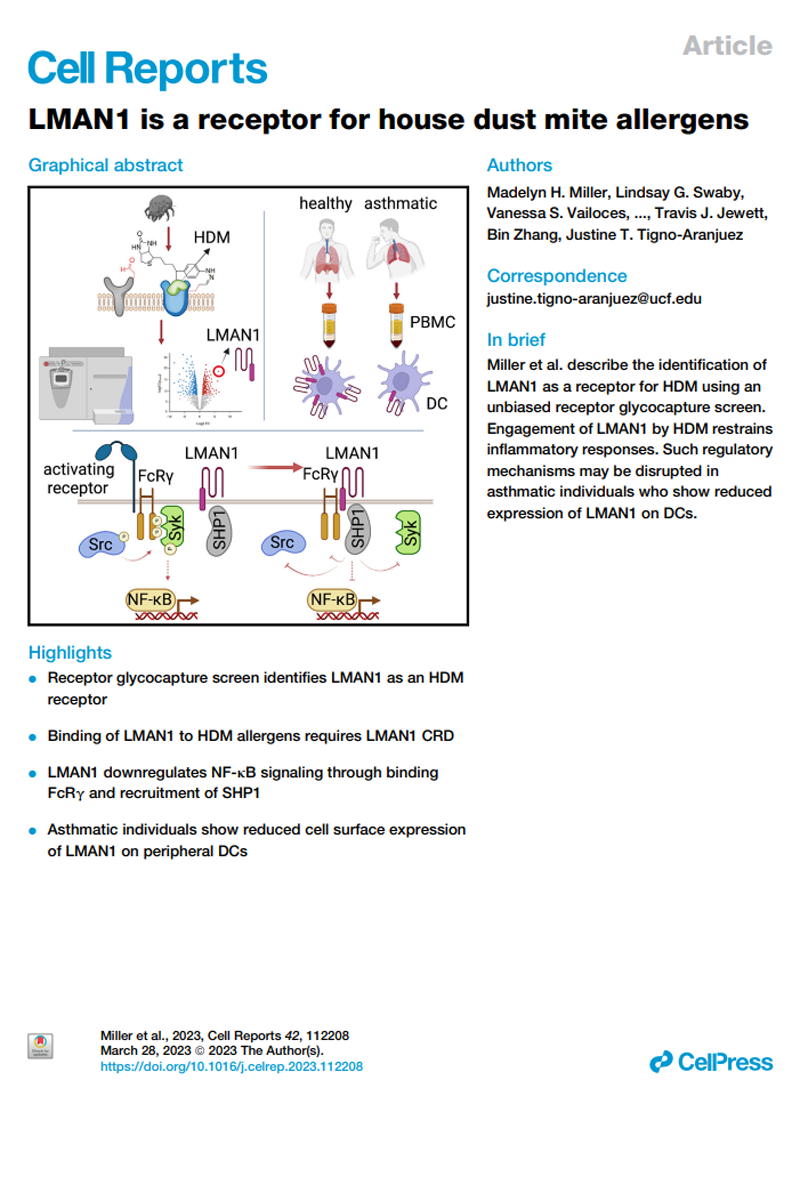
Identification of a receptor for house dust mite allergens
LRC-TriCEPS identified a completely new mechanism of monocyte activation that might be clinically relevant in Alzheimers’ and inflammation diseases. Feb 2023
Sensei Biotherapeutics Presents Preclinical Data at the 37th Society for Immunotherapy of Cancer (SITC) Annual Meeting
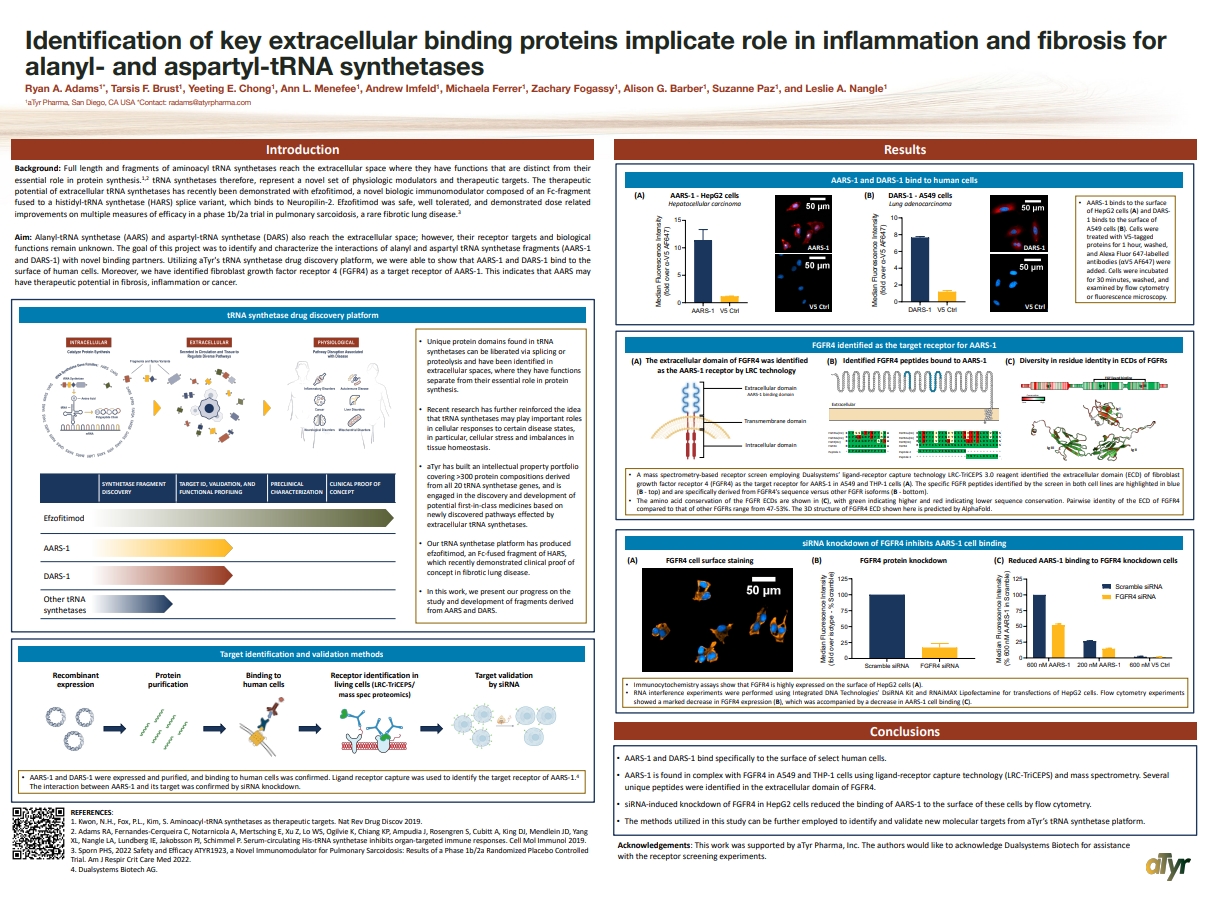
Identification of key extracellular binding proteins
Identification of key extracellular binding proteins implicate role in inflammation and fibrosis for alanyl- and aspartyl-tRNA synthetases
Latrophilin-2 is a novel receptor of LRG1
Latrophilin-2 is a novel receptor of LRG1 that rescues vascular and neurological abnormalities and restores diabetic erectile function – Diabetes mellitus (DM) is a chronic metabolic disorder that involves endothelial dysfunction and neuropathy and can lead to multiple complications, including cardiovascular disease, stroke, chronic kidney disease, foot ulcers, and retinopathy1. DM is also a major cause of erectile dysfunction (ED), which is a manifestation of microangiopathy and neuropathy1. Approximately 50–75% of male diabetic patients have ED2. Phosphodiesterase type 5 (PDE5) inhibitors are the most commonly used first-line treatment options for ED; however, these agents are ineffective in ~30% of patients and ultimately cannot rescue angiopathy and neuropathy in diabetic ED patients3. Tested alternative therapeutic options for ED include various angiogenic or neurotrophic factors, such as COMP-Ang1, vascular endothelial growth factor (VEGF), dickkopf2, neurotrophin-3 (NT3), and brain-derived neurotrophic factor (BDNF)4,5,6,7,8; however, these treatments have shown limited success in clinical trials. Experimental & Molecular Medicine volume 54, pages626–638 (2022)
Akkermansia muciniphila secretes a gluc.-like peptide-1-inducing protein that improves glucose homeostasis and ameliorates metabolic disease in mice
Nature Microbiol (2021) https://doi.org/10.1038/s41564-021-00880-5
Neuroplastin Modulates Anti-inflammatory Effects of MANF
iScience Volume 23, ISSUE 12, 101810, December 18, 2020, DOI:https://doi.org/10.1016/j.isci.2020.101810
NKG2A/CD94 Is a New Immune Receptor for HLA-G and Distinguishes Amino Acid Differences in the HLA-G Heavy Chain
International Journal of Molecular Sciences. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7352787/
Antimicrobial Peptides Derived from the Immune, Defense Protein CAP37 Inhibit TLR4 Activation by S100A9
METHODS. We used a TriCEPS-based, ligand-receptor glycocapture method to identify the
binding partners of CAP37 on live human corneal epithelial cells using the hTCEpi cell
line. We used an ELISA method to confirm binding with identified partners and test the
binding with CAP37-derived peptides. We used a reporter cell line to measure activation
of the identified membrane receptor by CAP37 and derived peptides.
Acidity changes immunology: a new VISTA pathway
Binding to carboxypeptidase M mediates protective effects of fibrinopeptide Bβ
https://doi.org/10.1016/j.trsl.2019.07.008, Translational Research Volume 213, Sörensen-Zender et al.
Nature Article VISTA interaction with PSGL-1 identified by LRC-TriCEPS
https://www.nature.com/articles/s41586-019-1674-5 Nature volume 574, pages565–570 (2019)
Anti-VISTA antibody that inhibits Vista function and blocks interaction with PSGL-1 and VSIG3 proteins slows tumor growth
https://www.nature.com/articles/s41598-020-71519-4 Mehta, N., Maddineni, S., Kelly, R.L. et al. An engineered antibody binds a distinct epitope and is a potent inhibitor of murine and human VISTA. Sci Rep 10, 15171 (2020). https://doi.org/10.1038/s41598-020-71519-4 PDF
Phage resistance at the cost of virulence
Phage resistance at the cost of virulence: Listeria monocytogenes serovar 4b requires galactosylated teichoic acids for InlBmediated invasion
PLOS Pathogens | https://doi.org/10.1371/journal.ppat.1008032 October 7, 2019
PDF
TFF3 interacts with LINGO2 to regulate EGFR activation for protection against colitis and gastrointestinal helminths
Nature Communications volume 10, Article number: 4408 (2019) https://doi.org/10.1038/s41467-019-12315-1
White Paper Ligand-Receptor Identification Methodologies Details Matter
European Biopharmaceutical Review April 2019, pages 16-20 / Maria P. Pavlou and Paul Helbling / White -Paper PDF
Validation of extracellular ligand–receptor interactions by Flow‑TriCEPS
BMC Research Notes 2018 11:863 https://doi.org/10.1186/s13104-018-3974-5 Laura A. Lopez‑Garcia, Levent Demiray, Sandra Ruch‑Marder, Ann‑Katrin Hopp, Michael O. Hottiger, Paul M. Helbling and Maria P. Pavlou Received: 28 October 2018 – Accepted: 30 November 2018- Published: 5 December 2018 PDF
Cardiac Targeting Peptide, a Novel Cardiac Vector: Studies in Bio-Distribution, Imaging Application, and Mechanism of Transduction
Maliha Zahid, Kyle S. Feldman, Gabriel Garcia-Borrero, Timothy N. Feinstein, Nicholas Pogodzinski, Xinxiu Xu, Raymond Yurko, Michael Czachowski , Yijen L. Wu, Neale S. Mason and CeciliaW. Lo Biomolecules 2018, 8, 147; doi:10.3390/biom8040147 Received: 24 September 2018 / Accepted: 8 November 2018 / Published: 14 November 2018
Leukocyte differentiation by histidine-rich glycoprotein/stanniocalcin-2 complex regulates murine glioma growth through modulation of anti-tumor immunity
Francis P Roche, Ilkka Pietilä, Hiroshi Kaito, Elisabet O Sjöström, Nadine Sobotzki, Oriol Noguer, Tor Persson Skare, Magnus Essand, Bernd Wollscheid, Michael Welsh and Lena Claesson-Welsh DOI: 10.1158/1535-7163.MCT-18-0097 Received January 27, 2018, Revision received April 21, 2018, Accepted June 19, 2018, Copyright ©2018, American Association for Cancer Research. PDF
Glycomics and Proteomics Approaches to Investigate Early Adenovirus–Host Cell Interactions
Lisa Lasswitz, Naresh Chandra, Niklas Arnberg, Gisa Gerold jmb Journal of Molecular Biology, doi.org/10.1016/j.jmb.2018.04.039 Received 15 February 2018, Revised 24 April 2018, Accepted 30 April 2018, Available online 7 May 2018.












 Dr Patric Delhanty
Dr Patric Delhanty



 (Quim) Madrenas, MD, PhD, FCAHS
(Quim) Madrenas, MD, PhD, FCAHS





 De'Broski R. Herbert Ph.D.
De'Broski R. Herbert Ph.D.