Scratching the surface
Technologies for targeting the cell surfaceome
HATRIC-LRC (Ligand Receptor Capture)
The TriCEPS / Hatric-LRC technology is exclusively available at Dualsystems. Please contact us to discuss your project.
The article was also published in the EBR Spring 2018. Please download here the white paper from Dualsystems
The development of methodologies that enable the study of the surface of a cell – the surfaceome – is pivotal to advancing the understanding of cellular differentiation, and through this will come the development of new, much-needed treatments.
The cell surface membrane – or plasma membrane (PM) – surrounds the cell providing necessary boundaries between the cytoplasm and the extracellular environment. This thin, semi-permeable membrane plays a vital role in protecting the integrity of the cell through selective movement of substances in and out. It also constitutes the base for the attachment of cytoskeleton and cell wall – for bacteria and plants – thereby providing and maintaining the shape of the cell. Moreover, PM allows cells to recognize one another and transmits signalling processes.
The building blocks of the cell membrane are lipids, proteins and their associated sugars. The composition and relative concentration of these molecules define the membrane function and vary among different organisms, cell types and cell states. Based on the fluid mosaic model introduced in 1972 by Singer and Nicolson, the PM is a mosaic of components – primarily phospholipids, cholesterol, proteins and associated carbohydrates – moving freely and fluidly in the plane of the membrane. Although it was thought that the distribution of components is uniform, current data suggest that the cell membrane is highly and tightly organised in heterogeneous microdomains: the maintenance of this heterogeneity is associated with a large energetic cost indicating its significance (1). In support of this hypothesis, perturbations to the lipid composition of the membrane that disrupt the proposed compartmentalisation drastically reduce the efficiency of signal transduction (1).
Introducing the surfaceome
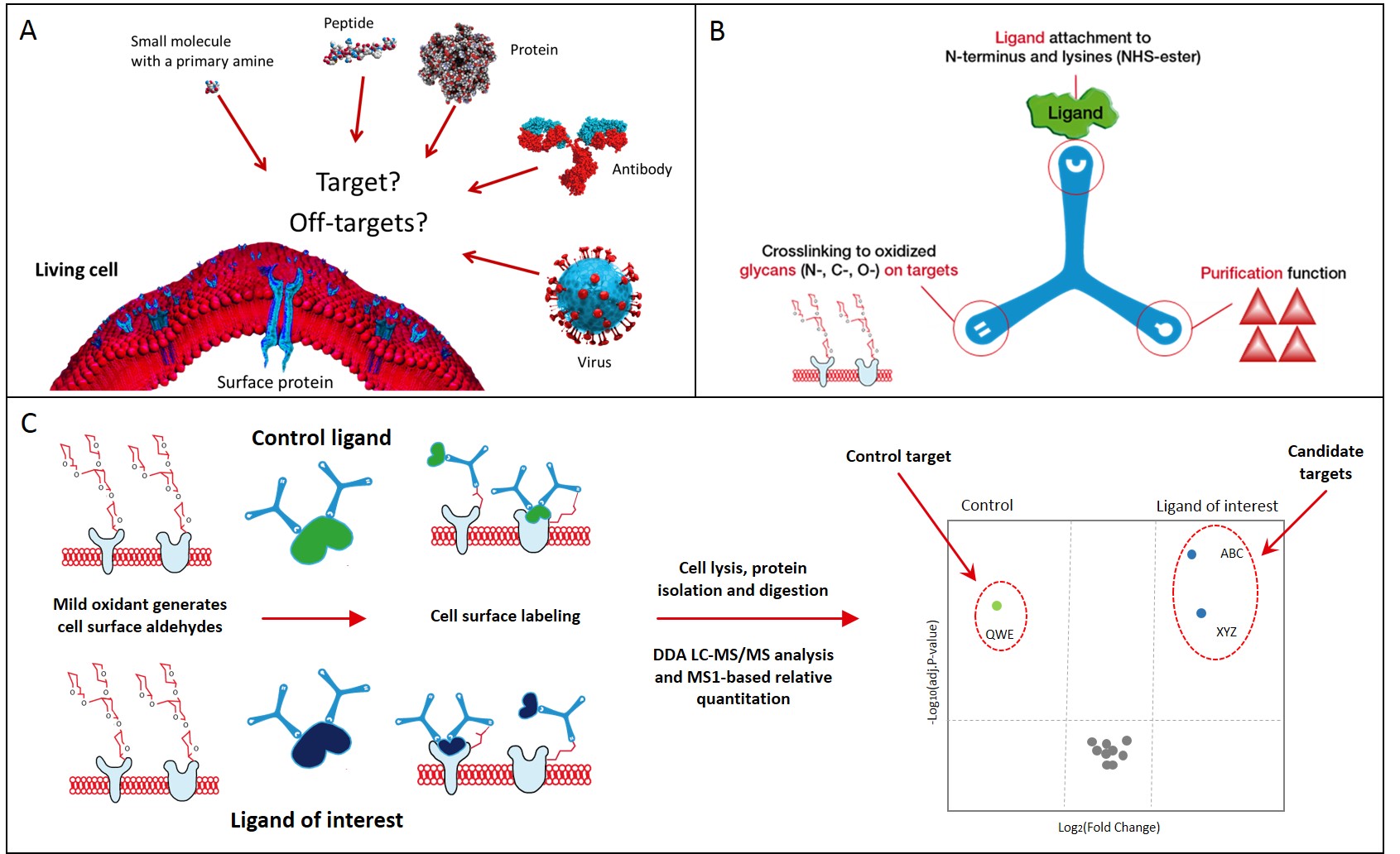
Although lipids and glycans are key components of the PM, the focus of the present review is on the collection of proteins that resides at the cell surface or surfaceome. Surface proteins can be physically embedded in the lipid bilayer (integral), be anchored to the phospholipids or integral proteins at either side of the cell membrane (peripheral) or even associate to the membrane only under specific conditions. The surfaceome constitutes roughly 50% of the PM mass (2) and exhibits a wide variety of functions. These include transport, enzymatic activity, signal transduction, cell-cell interaction and attachment to the cytoskeleton or the extracellular matrix. Different classes of surface proteins carry out these tasks for example channel and carrier proteins, enzymes, receptors, cell recognition and cell adhesion proteins. Given the range of functions carried out by surface proteins, it is not surprising that roughly 30% of predicted open reading frames in a typical genome encode membrane and PM proteins (2).
The surfaceome content differs among cell types and changes during developmental and disease states. Therefore, it contains unique markers that can be used to distinguish cellular phenotypes and disease states. These properties along with the fact that cell surface proteins are readily available make the surfaceome a rich source of phenotypic, diagnostic, prognostic and therapeutic targets that can be used in a variety of fields including oncology, immunology and stem cell research.
Overall, the development of methodologies that enable the study of the surfaceome is pivotal to advance our understanding regarding cellular differentiation and development, host-pathogen interactions and metastatic processes, and will lead to the development of new treatments.
Do you want to read more? Download the white paper.
TriCEPS compared to other techniques
| Technology for MoA studies for target identfication in the cell plasma membrane |
LRC-TriCEPS | Overexpression in HEK cells | Microarrays on artificial surfaces |
Pull down on cell lysates |
| On primary cells | Yes | No | No | Yes |
| On cell lines | Yes | Yes, but only HEK cells | No | Yes |
| No genetic modification of the cells | Yes | No | No | Yes |
| Cells are alive during interaction | Yes | No, cells are fixed | No | No |
| Targets are in the plasma membrane during interaction | Yes | Yes | No | No |
| Membrane proteins are correctly folded | Yes | Probably | No | No |
| Membrane proteins are correctly modified | Yes | Probably | Yes | Yes |
| Target proteins are in the correct microenvironment | Yes | Partly | No | No |
| No hypothesis for target interaction needed | Yes | No | No | Yes |
| Target proteins are immobilized | No | No | Yes | No |
| Target proteins are in the cell lysate during interaction | No | No | No | Yes |
| Targets are artificially overexpressed | No | Yes | No | No |
Download the white paper
We identify targets / off-targets of your ligand
If you want to get support for your project
Customers Testimonials – LRC-TriCEPS Service
Testimonials from our customers who have used the LRC-TriCEPS technology – in collaboration with Dualsystems Biotech AG.
University Hospital of Lausanne (CHUV)
Thanks to their LRC-TriCEPS platform, we were able to identify a cellular factor playing a major role in Zika virus entry. This unique method was applied to living cells without the need for genetic modification. In our case, the method worked robustly across three different cell types. Notably, it enabled the identification of an unexpected target at the cell surface that would likely have been missed by standard screening approaches, as the protein lacks a classical membrane anchor. What impressed us most was that the implication of this target, discovered via their screening, was subsequently confirmed in our in vitro studies using different cellular models.
We greatly appreciated the collaboration with the DualSystems Biotech AG team. Their expertise, responsiveness, and willingness to support our project at every stage were key to its success. The data provided were of high quality and well presented, facilitating downstream interpretation and validation.
Milos Stojanov, PhD - Head of the Materno-fetal and Obstetrics Research Unit - University Hospital of Lausanne (CH)

Miloš Stojanov, PhD | Senior Lecturer (MER1)
Head of the Materno-fetal and Obstetrics Research Unit
Department Woman-mother-child (DFME)
1011 Lausanne
Tél: +41 21 314 05 17
OncoLille Cancer Institute

Best,
Silvia Gaggero, PhD
Mitra Lab, Inserm
OncoLille Cancer Institute
Lille, France
AstraZeneca

James Dodgson
AstraZeneca
Cambridge, UK.
UCF College of Medicine
Justine Tigno-Aranjuez, Ph.D.
Assistant Professor of Medicine
UCF College of Medicine
Cohbar

Dr. Lindsay Stark
Drug Discovery Scientist at CohBar
Technical University of Munich
Using LRC-TriCEPS, we aimed to identify novel direct cell surface receptors of our ligand of interest.
At any time, we experienced great support of Dualsystems Biotech. They kindly helped to find optimal conditions for our purposes and provided help with any kind of question before, during and after the experiment. LRC-TriCEPS allowed us to identify novel cell surface receptors of our ligand, which we could successfully validate in different cell types and with different biochemical assays. We can fully recommend Dualsystems Biotech and are looking forward to perform further analyses using LRC-TriCEPS.

Prof. Dr. rer. nat. Achim Krüger
Institute of Experimental Oncology and Therapy Research
Klinikum rechts der Isar, Technical University of Munich
University of Miyazaki

Hideyuki Sakoda, MD, PhD
Associate professor
Department of Biological Sciences, Faculty of Medicine, University of Miyazaki, Japan.
Lund University Diabetes Centre

Dr. Claire L. Lyons,
Associate Researcher
Unit of Medical Protein Science
Lund University Diabetes Centre
Sweden
Australian National University

The Australian National University
Co-Director, Centre for Personalised Immunology, NHMRC Centre of Research Excellence
College of Health & Medicine
The Australian National University
Harvard Medical School, Brigham and Women’s Hospital


Maximillian Rogers, PhD
Research Scientist
Harvard Medical School, Brigham and Women's Hospital
Department of Medicine, Cardiovascular Division
Boston, MA
Center for Biomolecular & Cellular Structure, Institute for Basic Science

Associate Professor
Graduate School of Medical Science and Engineering, KAIST
Chief Investigator
Center for Biomolecular & Cellular Structure, Institute for Basic Science (IBS)
Department of Internal Medicine Erasmus MC
 Dr Patric Delhanty
Dr Patric DelhantyLaboratory of Metabolism and Reproduction
Department of Internal Medicine
Erasmus MC
Rotterdam, The Netherlands
Seoul National University

Chung Hwan Cho, Ph. D. candidate
Environmental Health Microbiology Laboratory
Department of Environmental Public Health
Seoul National University
Immuno-Oncology Discovery from Bristol-Myers Squibb published in Nature
Identification of a new immune-oncology drug target using the LRC-TriCEPS platform on primary human T-cells.
The University of Oklahoma – Health Sciences Center

Anne Kasus-Jacobi, PhD
Associate Professor of Research
University of Oklahoma Health Sciences Center
Department of Pharmaceutical Sciences
Oklahoma City, Oklahoma, USA
CuroNZ Ltd

Frank Sieg, PhD
CSO
CuroNZ Ltd
Mangawhai in New Zealand
University of Pittsburgh
Maliha Zahid, M.D., Ph.D.
Assistant Professor
Departement of Developmental Biology
University of Pittsburgh
University of Oklahoma Health Sciences Center

Anne Kasus-Jacobi, PhD
Assistant Professor of Research
University of Oklahoma Health Sciences Center
Department of Pharmaceutical Sciences
Oklahoma City, Oklahoma, USA
Biomedical Research Institute
The identification of a T cell co-receptor for staphylococcal superantigens had been challenging due to the structural features of the interaction and its kinetics. However, working with Dualstystems Biotech AG, and with Dr. Paul Helbling in particular, and using the LRC-TriCEPS technology, we were able to identify a candidate that was subsequently corroborated by biochemical and functional assays. We are very happy with this collaboration , and sincerely recommend it for the identification of novel receptor or co-receptor candidates.
 (Quim) Madrenas, MD, PhD, FCAHS
(Quim) Madrenas, MD, PhD, FCAHSChief Scientific Officer
Los Angeles Biomedical Research Institute at Harbor-UCLA Medical Center
Torrance, USA
QIMR Berghofer Medical Research Institute
 Anita Burgess | PhD
Anita Burgess | PhDHepatic Fibrosis Group
QIMR Berghofer Medical Research Institute, Australia
University of Miami, Miller School of Medicine
I would like to thank once again the company and, particularly, Dr Helbling for his attention and collaboration.
 Dr Karina Galoian
Dr Karina GaloianResearch associate professor
University of Miami, Miller School of Medicine
Department of Orthopedic surgery
Miami, Florida, USA
Münster University Hospital (UKM)
 Dr. rer. nat. Martin Herold
Dr. rer. nat. Martin HeroldWorking group from Prof. Dr. med. Luisa Klotz
Münster University Hospital (UKM), Germany
University of Manitoba
 Sari S. Hannila, PhD
Sari S. Hannila, PhDAssociate Professor, Department of Human Anatomy and Cell Science
Associate Member, Spinal Cord Research Centre
Max Rady College of Medicine, Rady Faculty of Health Sciences
University of Manitoba
The Rockefeller University
Assistant Professor of Clinical Investigation
The Rockefeller University
Medizinische Hochschule Hannover
East Tennessee State University
 Jonathan M Peterson
Jonathan M PetersonAssistant Professor
East Tennessee State University
Igenica Biotherapeutics
 Dr. Edward van der Horst,
Dr. Edward van der Horst,Senior Director, Preclinical Development
Igenica Biotherapeutics
Centro de Estudos de Doenças Crónicas
« The fruitful collaboration with Dualsystems Biotech using the LRC-TriCEPS (CaptiRec) technology showed that even on insect cells receptors could be identified »
Principal Investigator at CEDOC
Centro de Estudos de Doenças Crónicas
Washington University School of Medicine
University of California San Francisco
 De'Broski R. Herbert Ph.D.
De'Broski R. Herbert Ph.D.Assistant Professor in Residence
University of California San Francisco (UCSF)
Footer
Dualsystems Biotech AG
Dr. Paul Helbling
Grabenstrasse 11a
8952 Schlieren
Switzerland
phone +41 44 738 50 00
fax +41 44 738 50 05
LRC-TriCEPS Video
LRC-TriCEPS explained in a short video